Taxonomies for Natural History Collections
Presenting digital collections of natural history has its own unique set of challenges. This is something that Europeana's natural history aggregator OpenUp! knows well. With almost 9 million objects on Europeana Collections - visibility and search come with some very specific parameters. The below explores some of these key elements behind presenting natural history objects online and lets you know the best ways to find what you are looking for.

- Title:
- Botánica americana celestino mutis | CarlosVdeHabsburgo, Die im Bernstein befindlichen organischen Reste der Vorwelt, gesammelt (1845) | Neumann, bio bsu zoomus | Leony1, Cupboard2 MW Herbarium | Alexey Seregin, wikimedia commons, CC BY-SA
Natural history collections in museums worldwide are repositories of huge amounts of preserved biological specimens that document the past and present biodiversity of our planet, including many extinct species. Those collections contain objects like stuffed and mounted animals, pinned insects, dried plants, seeds, and fruit as well as all kinds of fossils. In the past, most of these specimens were accessible only to scientists, but today digitization makes them increasingly visible to the public. Virtual galleries of images and 3D models, as well as videos and audio files, allow exploring the hidden treasures of the museum’s depots that are usually off limits for regular visitors. Europeana’s natural history aggregator, OpenUp!, currently contributes 8.7 million objects from 34 institutions to the Europeana portal. That data provision relies on established data infrastructures in the natural history domain, namely the Biological Collection Access Service for Europe and the Global Biodiversity Information Facility.
![Ursus thibetanus G.[Baron] Cuvier, 1823, Museumfür Naturkunde Berlin, Germany, CC BY-SA](/files/Images/Blog_images/2019-01/Ursusthibetanus.jpeg)
Taxonomies represent our understanding of species biodiversity and evolution, which is subject to permanent research. Consequently, taxonomies are in constant flow. As new species are discovered, new names will be added. Systematic research might discover that a certain species is more closely related to another genus, so that part of the species’ binomial has to be changed. A genus might be merged with another genus or split up in several genera, which requires several species names to be changed. Whole taxon groups can be moved to other parts of the hierarchical tree as the result of newly discovered knowledge about common ancestry, e.g. in cases when traditionally used morphological characteristics have to be reconsidered in light of molecular evidence. Peculiarities such as homonyms (identical names for different species) and synonyms (several names for one species) add up to the difficulties dealing with taxonomies. The complexity of handling such dynamic data gave rise to the new field of taxonomic computing.
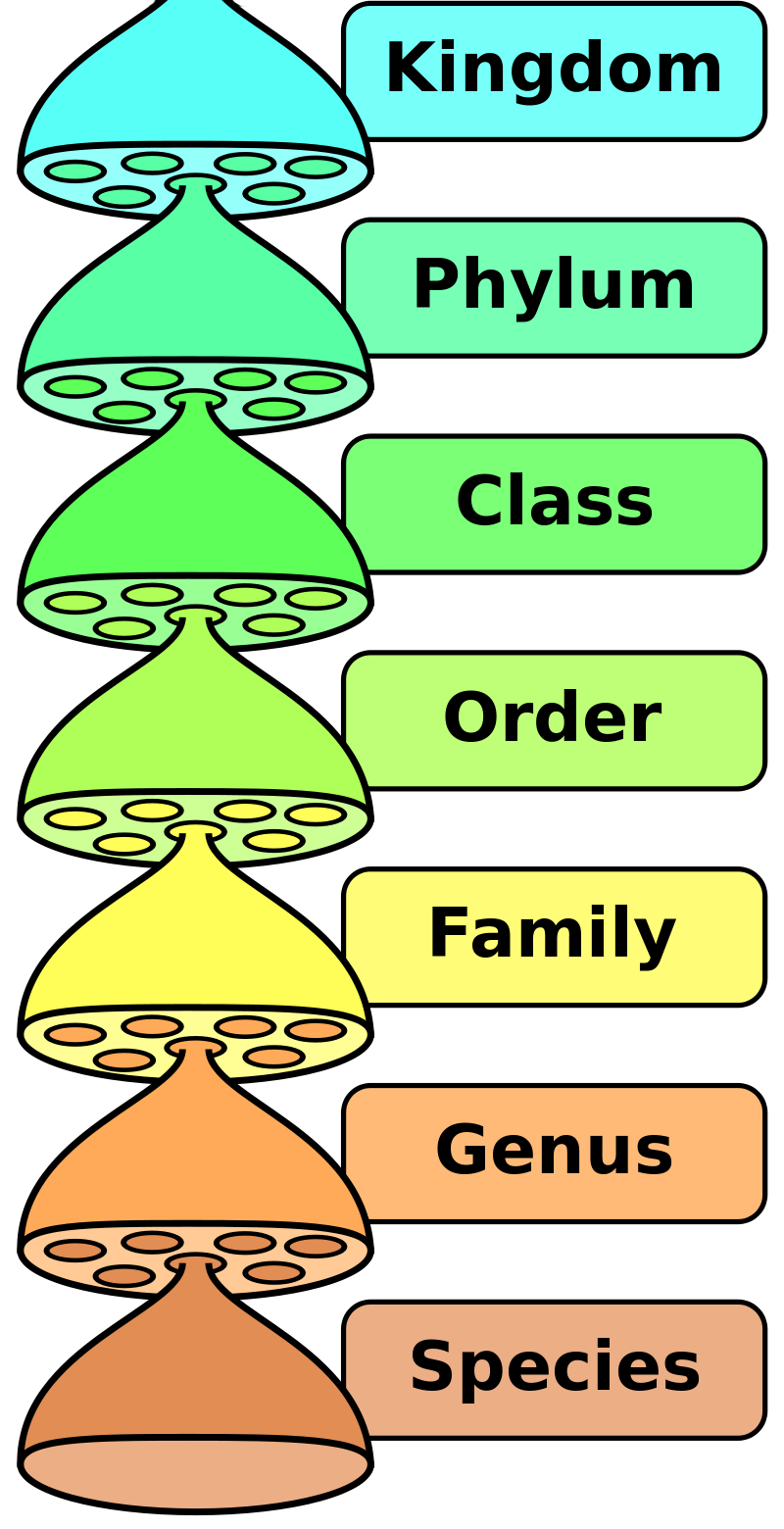
Traditional taxonomies often deal with a defined group of organisms, e.g. a certain family, class or kingdom, and refer to a certain geographic region, in which the described group is well-known and documented. Examples are regional ‘taxonomic checklists’ such as Euro + Med PlantBase (vascular plants of Europe and the Mediterranean region) and Fauna Europaea (European land and fresh-water animals), which are joint efforts of taxonomists from many institutions and are constantly being updated. Initiatives such as the Pan-European Species-directories Infrastructure (PESI) merge taxonomies from different communities into a single, all-taxa checklist. Similar initiatives exist on a global level: The Catalogue of Life pools data from 168 taxonomic databases into an authoritative index of known species of animals, plants, fungi and microorganisms, which currently lists 1.8m of the world's 1.9m named species. GBIF’s Backbone Taxonomy builds upon the Catalogue of Life and is regularly assembled in an automatic process from 56 sources.
Needless to say that the decision on which checklist should be used for a collection depends on the taxonomic and geographic coverage. Taxonomies undergo constant updates, so the matching of collection objects to any of the mentioned checklists should be performed at regular intervals. Most of them are available through web services that allow easy integration into existing infrastructures and products. Regional and global synonymised checklists such as PESI and the Catalogue of Life can be used to implement query expansion mechanisms that extend user queries for a taxon to all known synonyms of this taxon. Such query-expansion functions are already state-of-the-art in biodiversity portals.
For natural history specimens, Linked Open Data identifiers have become widely used in the recent past, for example by implementing the HTTP Stable Identifiers of the Consortium of European Taxonomic Facilities (CETAF). For taxa, similar initiatives are being discussed, but the inherent uncertainty and the constant flow of taxonomies make taxa not easy to grasp and hamper such efforts.
A problem that cannot be solved by using canonical taxonomies is the issue of misidentifications – specimens mistaken for a certain species, which results in incorrect names being used for objects. This cannot be completely avoided considering that some collections contain millions of specimens, which cannot be updated constantly. So this should be taken into account when using the data.
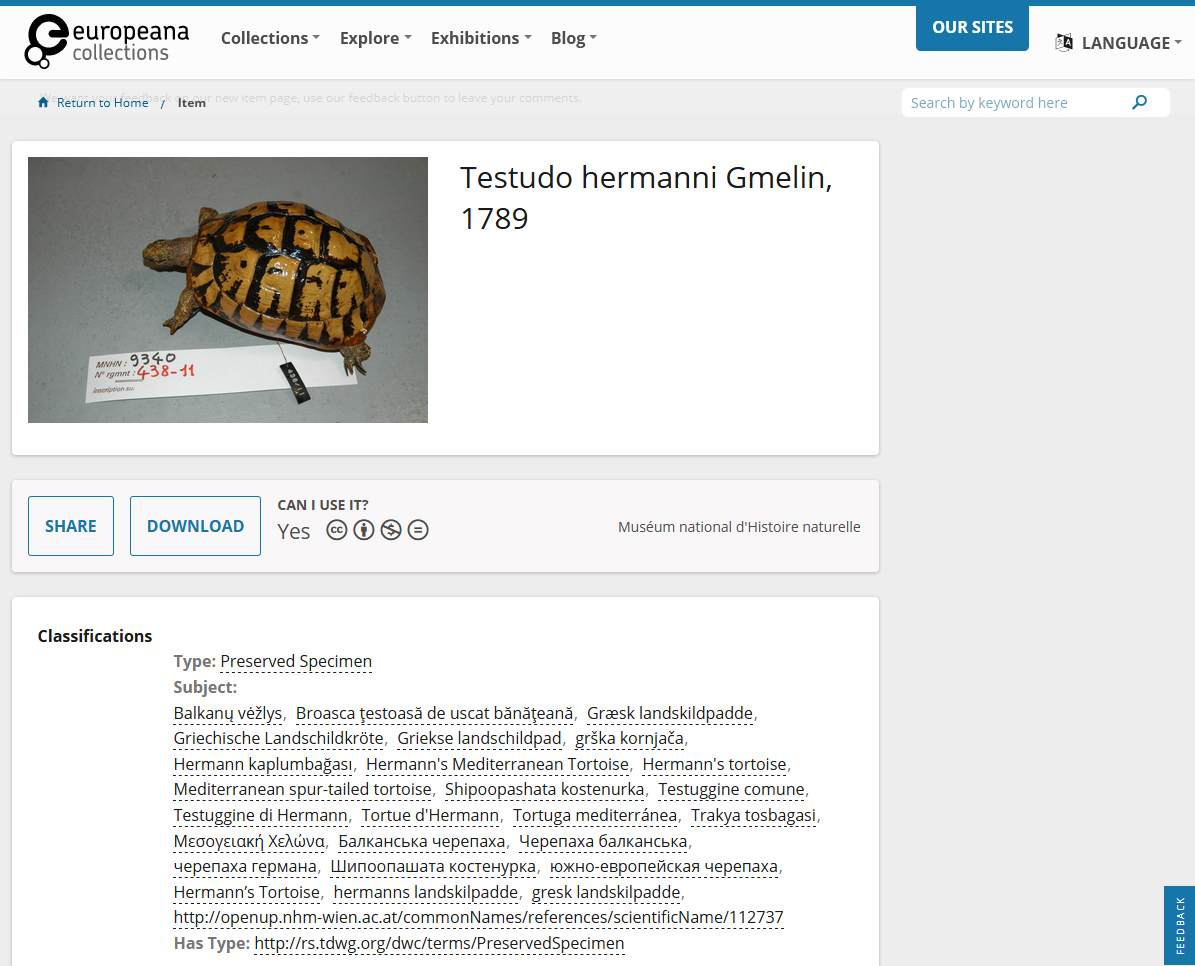
For OpenUp!, no uniform taxonomy is used for the specimen objects. As the data are provided by institutions that are experts in their respective fields, they are expected to apply appropriate checklists on their data before feeding them to OpenUp. However, to increase accessibility, OpenUp! enriches the objects’ metadata with common names in 300 languages and dialects, so that a species can be found (with some certainty) without knowing its scientific name. Further enrichment includes links to scientific literature available in the Biodiversity Heritage Library (BHL), a consortium dedicated to making legacy literature on biodiversity accessible online.
Acknowledgements: I’d like to thank my colleagues Walter Berendsohn, Petra Böttinger, Gabi Dröge, Anton Güntsch, Agnes Kirchhoff and Gerda Koch for their valuable comments and suggestions.
Image attributions:
- Ursus thibetanus G.[Baron] Cuvier, 1823, Museumfür Naturkunde Berlin, Germany, CC BY-SA
- A biological classification’s seven major taxonomic ranks, Peter Halasz, Wikimedia Commons, Public Domain.
- Testudo hermanni Gmelin, 1789, Muséum national d'Histoire naturelle, France, CC BY-NC-ND
